Targets
Note about BayBE
Catalyst uses BayBE under the hood. BayBE may evolve over time. This page explains parameters in non-technical terms as they are used in Catalyst today. For the detailed technical reference that Catalyst is aligned with, see the BayBE 0.13.2
Experiments in Catalyst use targets to describe what "good" looks like. A target is the outcome you measure after each trial (for example: yield, impurity, viscosity, cycle time). BayBE learns from your trials and can recommend what to run next.
Where you see targets in Catalyst
In the Create Experiment wizard, targets are configured in the Target step.
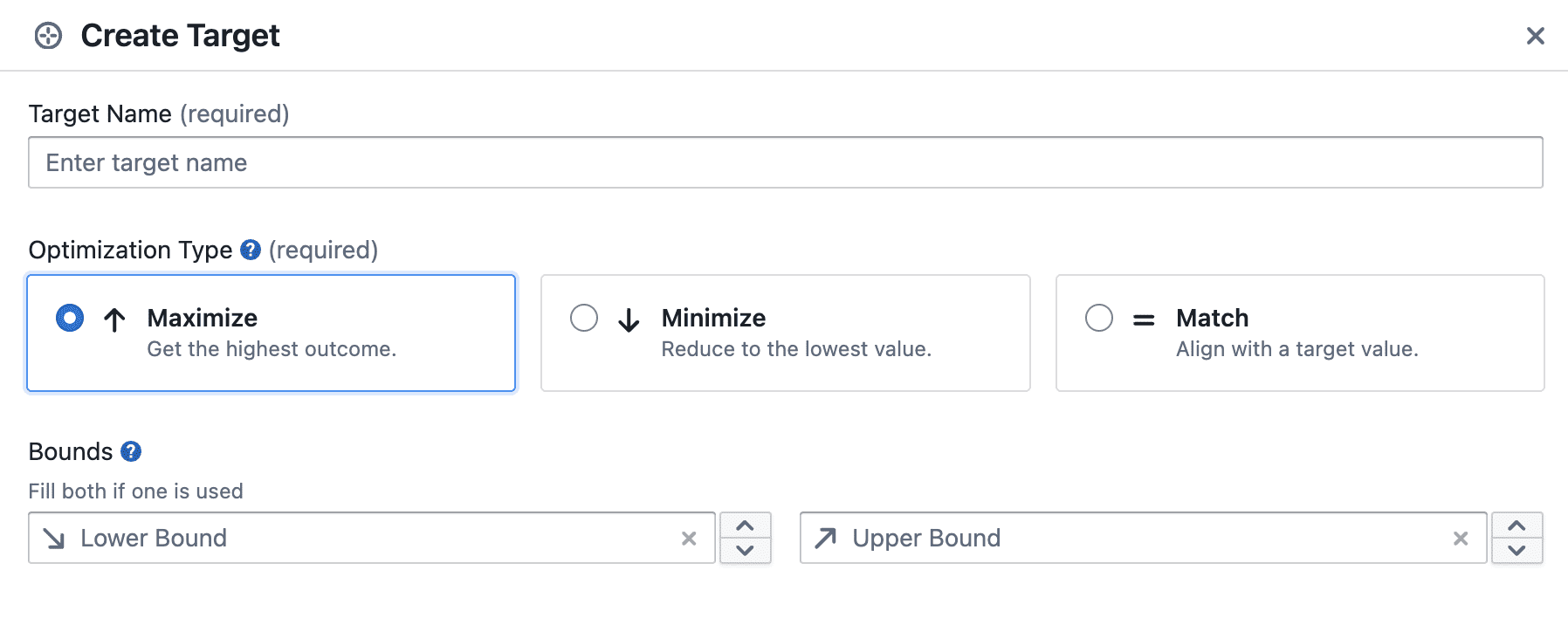
In this step you typically:
- Add one or more targets (what you will measure per trial)
- Choose the direction for each target (maximize / minimize / match)
- Optionally set bounds (lower and upper) for a target
- (If your UI supports it) choose how multiple targets are combined into one overall goal (objective/scalarization)
What makes the Target step "invalid"
Your UI should block progress when:
- No targets are defined
- Any target has validation errors
- Any target has bounds where lower bound ≥ upper bound (must be strictly lower)
This keeps the optimization problem well-defined.
What is a target?
A target is an output you care about. It could be something you want to:
- Maximize (bigger is better)
- Minimize (smaller is better)
- Match a value (best is "close to a desired value") — BayBE supports target matching.
Everyday analogy (for intuition)
If you are baking a cake, targets might be:
- Taste score from tasters — maximize
- Moisture level — maximize
- Baking time — minimize, but only as long as the cake is baked through
For coffee, targets might be:
- Bitterness score — minimize
- Strength — match a preferred value (e.g. "around 7/10")
These analogies map directly to more realistic process targets like yield, impurity, and viscosity.
Practical examples
For a reaction or process experiment, targets might include:
- Yield (%) — maximize
- Impurity (ppm) — minimize
- Viscosity (Pa·s) — match a preferred operating value (stay near a target range)
You can:
- Choose one main target (for example, maximize yield), or
- Combine several targets into a single overall score (for example, a "desirability" / scalarization approach), or
- Look at trade-offs (Pareto-style) where your workflow supports it.
One target vs multiple targets
Single target (most common)
You pick one main outcome (e.g., "maximize yield"). This is the simplest setup.
Multiple targets
Sometimes you care about multiple outcomes (e.g., maximize yield and minimize impurity, while keeping viscosity in a range).
User-friendly ways to think about it:
- Trade-off view (Pareto): compare options that represent different trade-offs
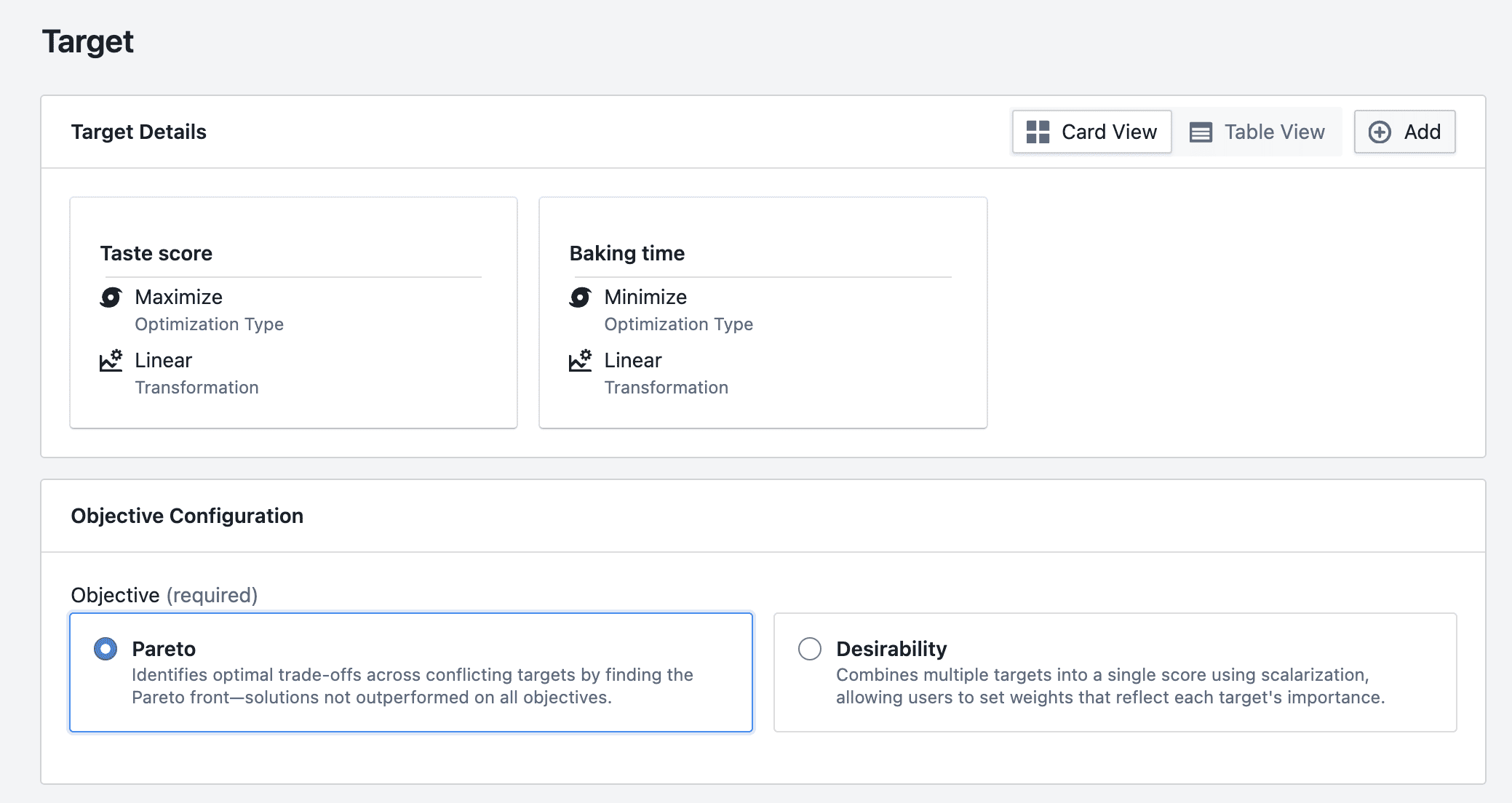
- Combine targets into one overall score (often called "desirability" / scalarization)
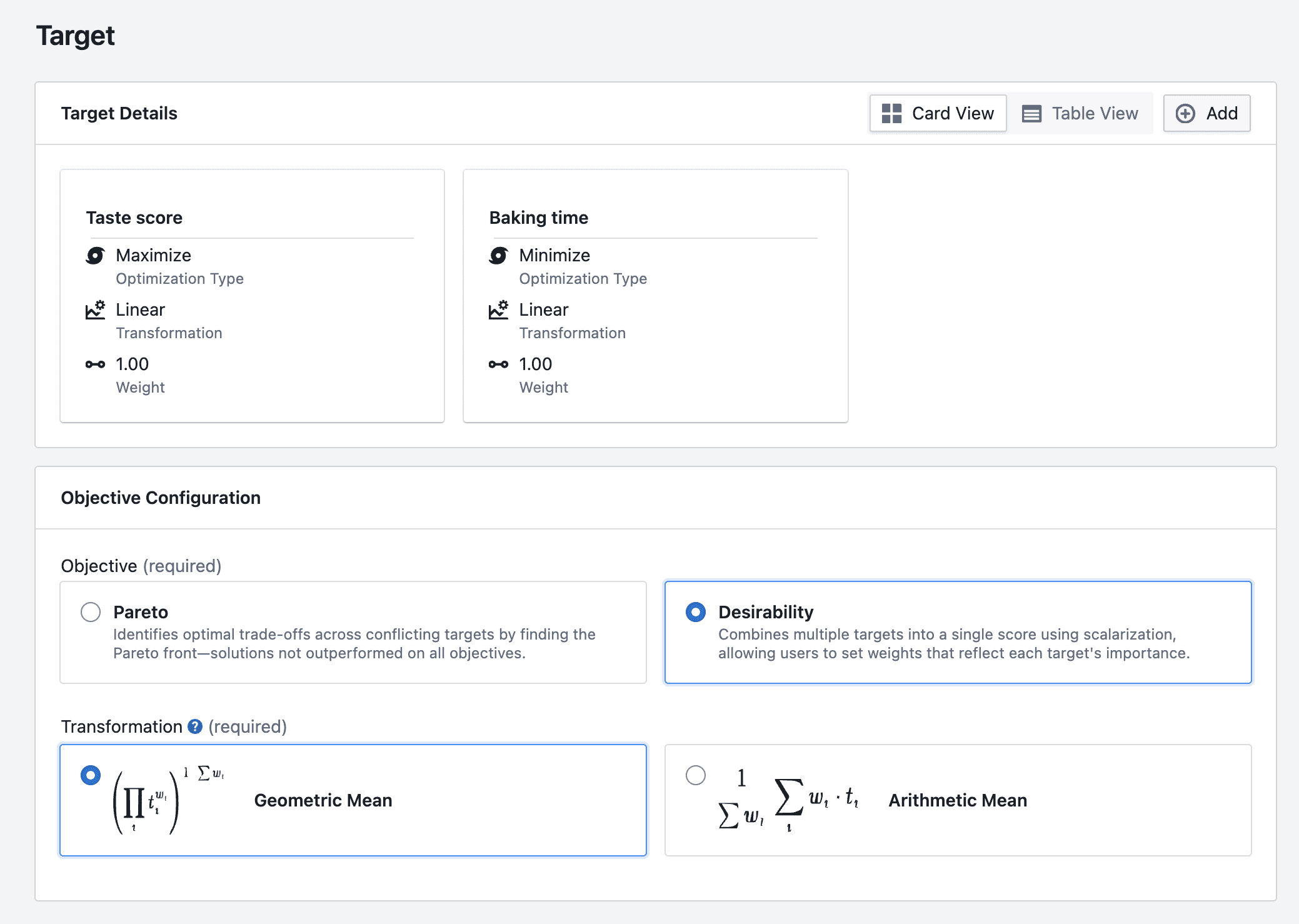
BayBE supports multi-target approaches such as desirability scalarization and Pareto optimization.
Bounds (optional) vs constraints (don’t confuse them)
- Target bounds describe a valid/expected range for the measured outcome or help define how "good" is computed (for example, what "too low" or "too high" means).
- Constraints restrict which parameter combinations are allowed.
If you allow bounds in the UI:
- If both are set: lower < upper (strict)
Targets and noise
Real measurements are noisy. BayBE is designed to handle variation across trials.
If your process is very noisy:
- Run replicates (same parameters multiple times)
- Record the average (and optionally variability) as the target value
Practical tips
- Choose targets that truly represent success (for example, final yield, not an intermediate proxy, unless that proxy is what you actually care about).
- Keep target definitions consistent during an experiment; if the measurement method changes, consider starting a new experiment.
- Record units and calculation method (e.g., "purity by HPLC, % area").
Further reading
- BayBE Targets: https://emdgroup.github.io/baybe/0.13.2/userguide/targets.html
- BayBE Objectives: https://emdgroup.github.io/baybe/0.13.2/userguide/objectives.html